HOME
>
RESEARCH DETAIL
Chameleon Emulsion (in Japanese)
From recent publicatons
Synthtic DNA nanounits can develop submicron-sized, single-molecule organisate
Structures made by DNA are attracting a lot of attention. For
example, DNA oligomers fold long single-stranded DNA, which serves as a template, into arbitrary shapes by combining short strands of DNA (stapled DNA) that are complementary to specific sequences in the DNA. Our research focuses on the characteristic branching structures (replication forks or Holliday junctions) that occur during the DNA replication process. Here, we focused on the phenomenon of self-organized association of DNA double helices with trigeminal structures. However, since the association of DNA is reversible, it dissociates at high temperatures or in solutions with low ionic strength. Therefore, we combined it with a low molecular weight compound (psolaren) that can cross-link DNA strands by generating covalent bonds through photoreaction. As a result, we discovered that the DNA can grow to submicron size by generating hyperbranched polymers on the substrate surface. It is expected to be used as a single molecule electronic material in the future.

From recent publications
Analyze pesticide residues using antigen-antibody reactions.
Organisms try to eliminate foreign substances, such as
infectious pathogens, by producing antibodies. Antibodies precisely recognize and bind to foreign substances, so they can be artificially produced for a variety of uses. The enzyme immunoassay is a typical example of such a method, which enables highly sensitive and selective analysis. We have been conducting research on a new analytical system, the microfluidic analysis system, which integrates a series of analytical operations, such as sample introduction, separation, and detection, into a single palm-sized substrate. This time, we have established a system that incorporates enzyme immunoassay and developed a high-performance method for the analysis of pesticide residues. Using this system, there is no need for expensive and large analyzers. It can be brought to the site where analysis is required and measured on the spot. Therefore, it is expected to be applied to environmental analysis in the future.

Electrochemiluminescence : ECL
We employ electrochemical reactions that emit fluorescence with high efficiency.
UV-vis absorption as well as fluorescence spectrometers
tend to be complex and large instruments. On the other hand, electrochemiluminescence spectrometry is compact and suitable for microfluidic analysis systems, but its detection sensitivity is not as good as that of fluorescence. Therefore, we are studying microanalytical systems incorporating electrochemiluminescence analysis, which combines the advantages of both methods. In addition to studying the conditions for efficient reactions, we are also working on the development of compact, high-performance detection devices in conjunction with research on the synthesis of our own luminescent materials.
Spectroelectrochemical Analysis Method
Analyze unstable and short-lived chemical species with high accuracy.
Analytical methods for unstable or short-lived chemical species,
such as radicals, have not been developed so far. Therefore, we have started to apply a multi-channel spectroscopic system to model radical species generated by electrode reactions. In addition to measuring the absorption spectra of the chemical species generated at the electrode/solution interface with high speed and high sensitivity, we are applying our own analysis method and are gathering interesting data.

Carbon Quantum Dots:CQD
We apply "green" quantum dots to bioanalysis.
Quantum dots (CDs), which are inorganic semiconductors, are
fluorescent nanomaterials that have features not found in organic compounds, but the fact that they contain toxic heavy metals has been a bottleneck in their application. Recently, nanoparticles made almost entirely of carbonaceous materials have been reported. Carbon quantum dots (CQDs) are beginning to be used in the biotechnology field, not only because of their unique fluorescent properties, but also because of their low toxicity to living organisms. We are the first to report that CQDs exhibit good properties as fluorophores in peroxalate chemiluminescence analysis. Research on environmental analysis methods using CQD has been engaged
Single Molecular Imaging
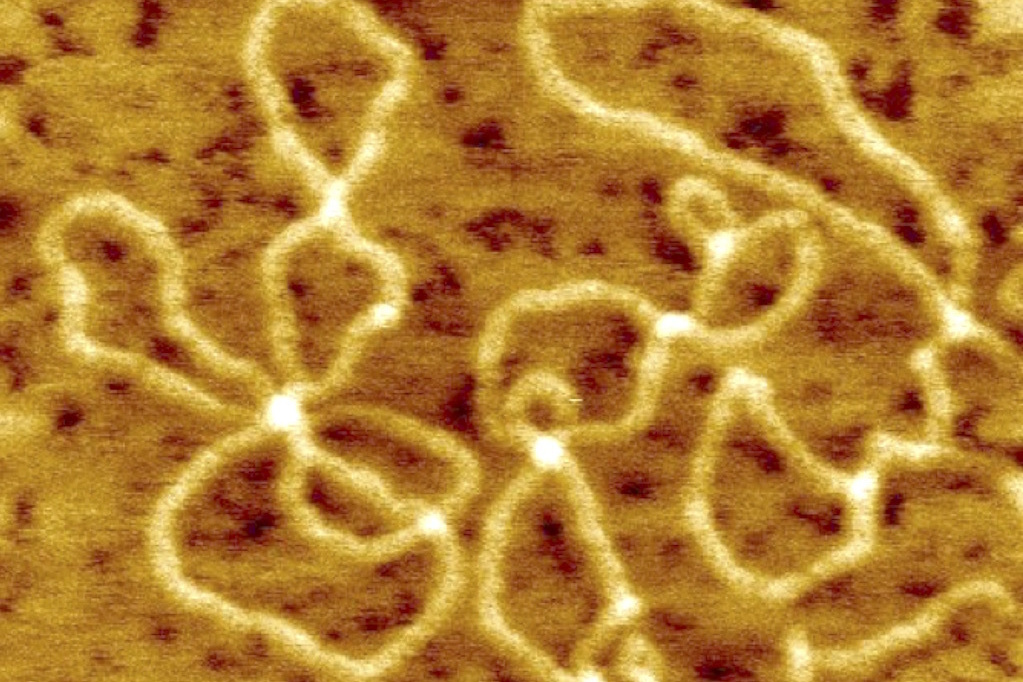
Analyze biomolecules based on single-molecular imaging using an atomic force microscope.
Shown is an example of molecular image of pBR322 DNA
obtained by atomic force microscope. The plasmid's unique super-helical structure (twisted DNA double helix. The image of a twisted rubber band) can be clearly seen at the level of a single molecule. We have designed the probe DNA sequence so that the supramolecular structure changes dramatically when the target DNA binds to it. By doing so, we proposed an analytical method to detect the target DNA by single molecule observation using AFM. In addition to this, single molecule observation of enzymes and proteins is possible, and we are studying the visual detection of enzyme reactions.
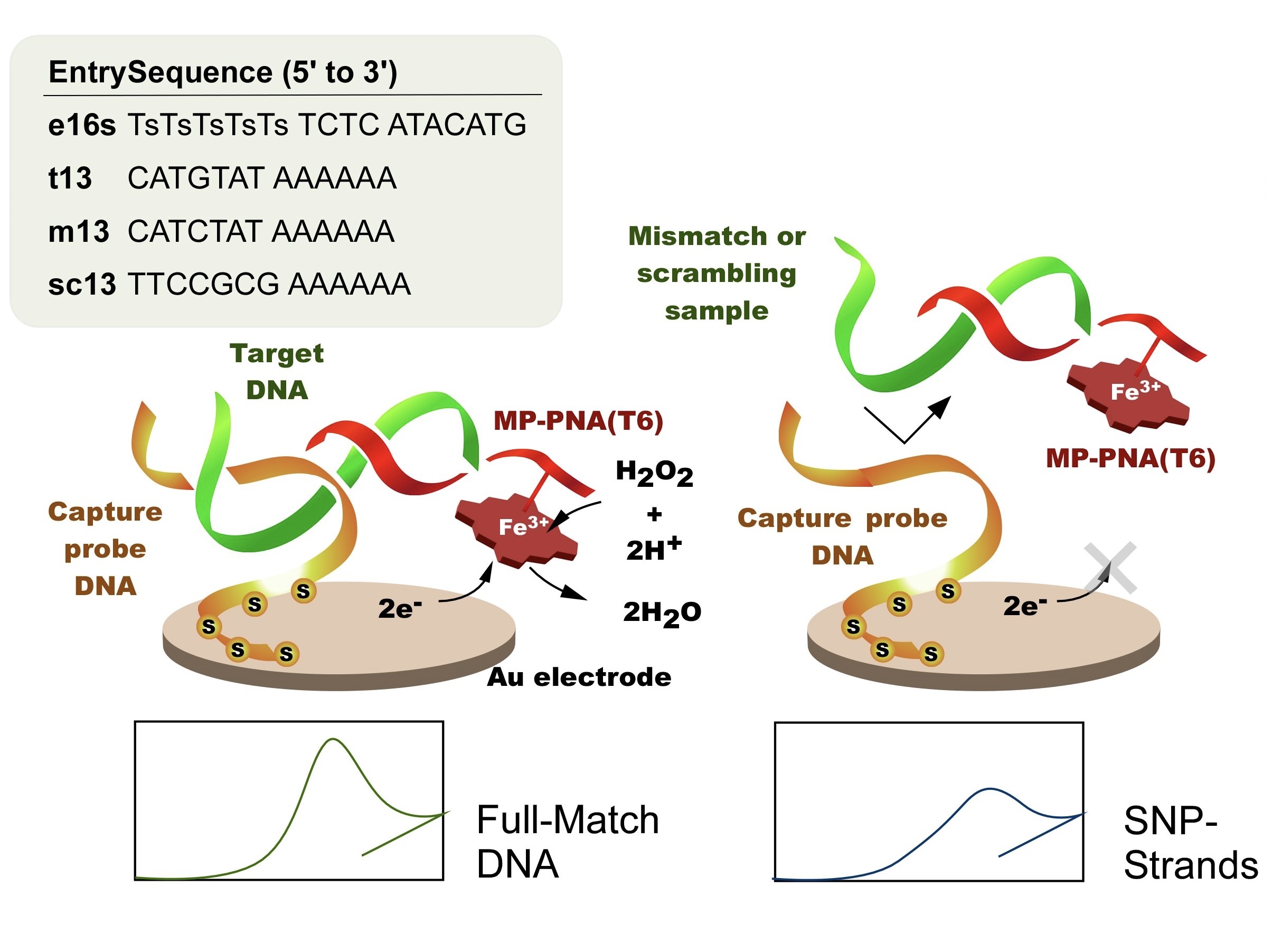
DNA Sensor/Gene Sensor
Detects DNA-binding drugs and mutations of genes by electrochemical measurement.
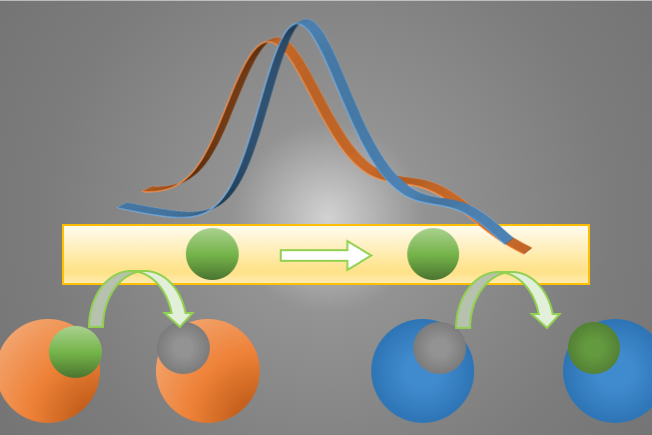
Target DNA is selectively derivatized into a sandwich complex
including probe DNA and labeled DNA for electrochemical detection. DNAs do not exert any catalytic reaction that are observed in enzymes. Moreover, the guanine bases are easily degraded by redox reactions, making it difficult to detect using common assays in biosensors. Here, the target DNA contains a genetically meaningful sequence (gene) and an associated sequence. We have devised a method in which a third DNA fragment is labeled with a redox-active small molecule compound in addition to the DNA fragment used to recognize the former (DNA probe). If the sequence is complementary to the sequence associated with the gene, a hybridization product consisting of three DNA strands is generated, as shown in the figure, and the target DNA can be detected electrochemically. If the target DNA contains a single nucleotide polymorphism, the measurement conditions could be set so that hybridization does not occur.
PNAzyme
This is a monolithic nucleic-acid and peptide-enzyme conjugate exerts both function.
Soon after the structure of the DNA double helix was revealed
by Watson and Crick (1953), research on artificial nucleic acids or nucleic acid analogs began. Peptide nucleic acids (PNAs) are one such type of nucleic acid, in which nucleobases are linked via amide bonds, the same as in peptides, instead of the phosphate diester bonds found in nucleic acids. On the other hand, as seen in ribozymes, nucleic acids are often involved in biological phenomena by expressing enzymatic functions. This deviates from the route of enzyme production by transcription of nucleic acids, which has led to a discussion comparing the relationship between nucleic acids and enzymes to that of an egg and a chicken. Focusing on the structure of PNA, we have proposed an artificial nucleic acid catalyst, PNAzyme, which is synthesized by integrating a catalytic peptide. We have succeeded in synthesizing a PNAzyme that exhibits peroxidase activity, and have reported its application to bioanalysis.


Designer Protein
This is an artificial protein assembled from functiona unit structures.
We are working on artificial proteins designed by assembling
domains responsible for molecular recognition and catalysis. Enzymes, proteins, and receptors express their functions by adopting a special three-dimensional structure. In other words, peptides with specific amino acid sequences are arranged in domains that maintain secondary structures such as helices and sheets, and are responsible for molecular recognition and catalysis. We are challenging the realization of artificial protein enzymes by extracting the basic functional units and reproducing the spatial structure by taking advantage of solid-phase peptide synthesis.